23andMe’s Paternal Haplogroup Report describes the genetic line connecting you to your father, his father, and beyond. This blog post describes some of the science behind the report and the details of the update to our paternal haplogroup assignments to improve accuracy.
Overview
To identify the paternal haplogroup of each customer with a Y chromosome, 23andMe uses our “Yhaplo” program, which relies on Y-chromosome variation data curated by the International Society of Genetic Genealogy (ISOGG). In the years since we first developed Yhaplo, ISOGG has updated various entries, leading to incorrect (but closely related) paternal haplogroup assignments for some customers.
We have identified and corrected several “variant metadata” errors for this update to improve haplogroup assignments. While we were at it, we also excluded variants performing poorly on the latest genotyping chip, V5, further improving haplogroup calls. As a result, some customers will see enhanced paternal haplogroup assignments, whereas most will not.
Scientific Details
Haplogroups and Haplogroup Nomenclature
The Y chromosome bears the longest stretch of non-recombining DNA in the human genome, making it a powerful genealogical tool. The chromosome contains sufficient information to reconstruct a detailed phylogenetic tree relating the male lines of every man to the most recent male-line ancestor of all men. The clades of this tree—the subtrees descending from any given branch—are called “haplogroups.” Because haplogroups are highly correlated with geography and population, they can tell us where an individual’s male-line ancestors may have lived and give us insight into historical migrations.
The Y-chromosome tree in Figure 1 illustrates the primary structure and indicates how the major haplogroups relate. Each branch shown is the root of a subtree with a far more detailed structure not shown.
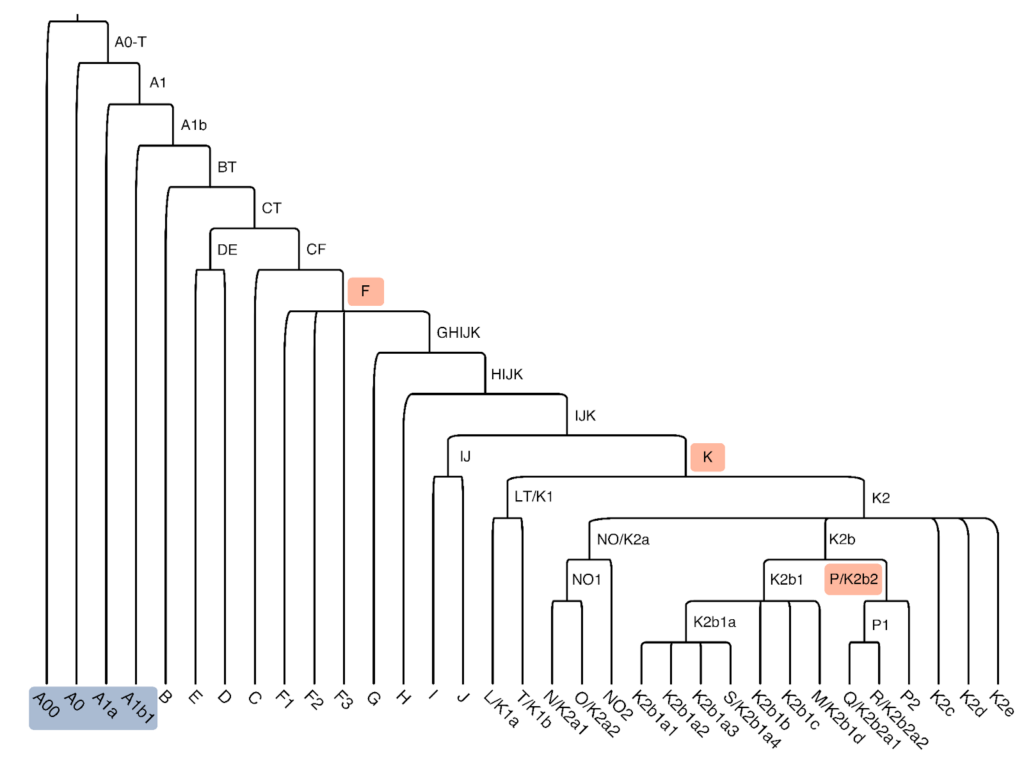
Every tree branch represents a set of one or more genetic variations, each of which arose in some ancestor of the clade. For example, on the Y chromosome of an individual who lived ~35,000 years ago, an adenine (A) mutated to a guanine (G) at GRCh37 position 15,581,983. Today, this individual has many male-line descendants, all of whom carry the “derived” G allele at this genomic position. In contrast, men who do not count this individual among their male-line ancestors usually carry the “ancestral” A allele.
When a variation arises, we call it a “mutation,” and when it has risen to an appreciable frequency, we call it a “polymorphism.” The single nucleotide polymorphism (SNP) described above is “M207”. The “M” stands for “marker”, and the number indicates that it was the 207th Y-chromosome marker discovered by Peter Underhill, who named the SNPs he discovered in sequence with “M” numbers. There are many other phylogenetically equivalent SNPs on the branch defining haplogroup R. Still, since M207 is well known, we use it as a proxy for the others and refer to the haplogroup as “R-M207.” In general, we refer to each haplogroup by one or more letters, a hyphen, and the name of a SNP associated with the branch defining the haplogroup.
Identifying an Individual’s Y-Chromosome Haplogroup
Yhaplo walks along the tree to determine how a customer’s Y chromosome relates to the known Y-haplogroup phylogeny.
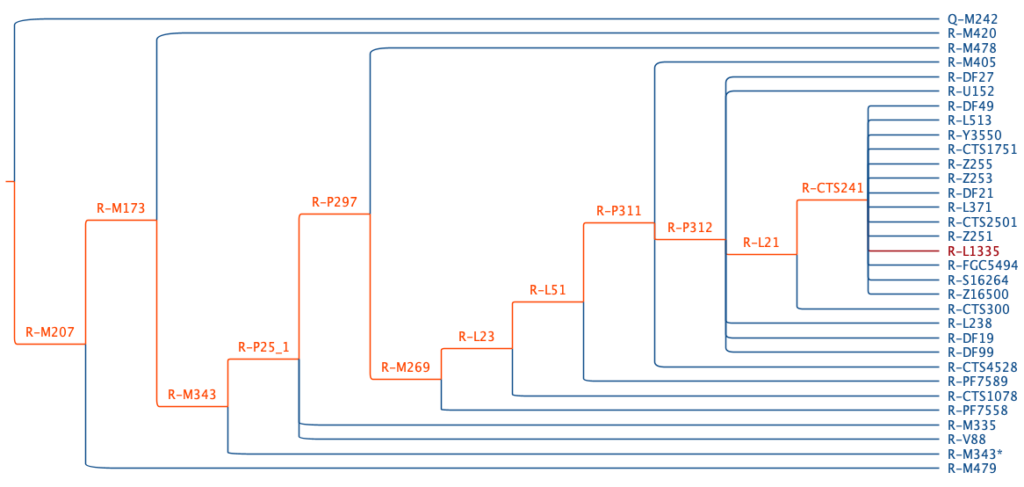
Each considered branch compares the individual’s observed genotypes to the ancestral and derived alleles of markers associated with the branch. It decides whether to consider the branch’s descendants or to move on. In doing so, it traces a path from the tree’s root to a final haplogroup designation. Figure 2 shows an example. In this example, an individual possesses derived alleles along a path (orange branches). This extends from the tree’s root (not shown) to the root of haplogroup R (R-M207) to haplogroup R-CTS241. In contrast, the individual possesses ancestral alleles (blue branches) outside this path.
Why Some Haplogroup Assignments Have Changed
As described above, Yhaplo draws variant metadata from ISOGG. This includes the ancestral and derived alleles of each variant. For most people, sporadic metadata errors did not impact Yhaplo’s ability to call haplogroups. However, in some cases, they led to haplogroup assignments that were a bit off. For example, the ancestral and derived alleles of SNP L1335 were reversed. This led to Yhaplo incorrectly assigning R-L1335 to some individuals (red branch in Figure 2). With such errors corrected, Yhaplo is more accurate, and these changes will be reflected in the updated Paternal Haplogroup Report.
Additional Resources
For additional details on the Yhaplo algorithm, please see our bioRxiv manuscript. We have open-sourced Yhaplo on GitHub for non-commercial use, pursuant to the terms of the non-exclusive license agreement included with the software distribution.
Learn More
You can read more about haplogroups in this blog post, or look at some of the frequently asked questions below. Find out more about 23andMe’s Ancestry Service here.
What’s a haplogroup? Scientists use the term haplogroup to describe a group of mitochondrial or Y-chromosome sequences that are more closely related than other sequences. The term haplogroup is a combination of haplotype and group. In this context, haplotype refers either to the DNA sequence of one’s mitochondrial DNA, inherited from one’s mother or to the DNA sequence of one’s Y chromosome, passed from fathers to their sons. Haplogroups are assigned by detecting specific genetic variants unique to each haplogroup.
What’s a maternal haplogroup? Your Maternal Haplogroup Report tells you about your maternal-line ancestors, from your mother through her mother and beyond. If
If you are male, your Paternal Haplogroup Report tells you about your paternal-line ancestors, from your father to his father and beyond.
What might my haplogroup tell me about my ancestry? Your haplogroup is a clue to your maternal or paternal ancestry. Over tens of thousands of years, humans migrated from eastern Africa to inhabit every continent on Earth except Antarctica. The Haplogroup reports show the migration patterns of people with a given haplogroup.