The sobering fact about drug development is that it takes too long and costs too much money. 23andMe wants to change all that.
At the annual meeting of the American Society of Human Genetics this month, 23andMe computational biologist Fah Sathirapongsasuti offered some insight into how this is being done.
“It’s a well-appreciated fact among drug developers that genetic evidence can indicate success of a drug program, but until now, genetic data was hard to come by.”
In his presentation, Fah explained 23andMe’s unique drug discovery model, which has the promise of lowering the cost and accelerating the pace of drug discovery. This new model is also what is behind 23andMe’s Therapeutics group.
The current industry model isn’t working well, he said. Nine out of ten drugs deemed successful in animal studies fail in human clinical trials. And all that time and all those failures add up to staggering costs. Although there’s a dispute about just how many hundreds of millions or billions it costs to develop a drug, no one disputes that finding new drugs is too slow and costs way too much.
But there’s another way, as Fah explained.
Simply using genome wide association studies can help improve success.
A recently published study showed that starting with a GWAS could double the success rate of drug development. Another approach is to identify genetic variants that are protective and use those variants to zero in on promising drug targets. A well-known example of that approach is the work done by Amgen and Sanofi in the development of two recently approved drugs that target cholesterol-lowering variants in the gene PCSK9, Fah said.
But what makes 23andMe research model unique is its customers. So far we have more than a million, and 80 percent of them have chosen to participate in our research. They are also engaged and have shared more than 300 million data points about themselves that inform research. Our scientists can gather that information about health history, or drug sensitivity or ancestry and traits, into specific “phenotypes.”
We have more than a thousand phenotypes to work with such as phenotypes for cardiovascular and autoimmune diseases, as well as different forms of cancer and metabolic conditions, giving our researchers a tremendous resource to compare genetic information against.
That gives 23andMe researchers a huge head start, said Fah. Using that data, researchers first look at phenotypic information to find a candidate indication for genes that, in turn, can be easily targeted by antibodies. 23andMe recently did just that with, studying 255 genes that code for cytokines and receptors, and focusing on those genes that have not yet been targeted by industry. About half of that total have not yet been used for drug targeting.
Finally screening those targetable variants in our database, our researchers found about two-dozen potential targets.
There’s a lot of additional work that goes into the process, for instance just determining whether these targets offer a plausible biological pathway that’s relevant to treating a disease.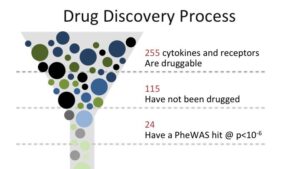
Fah illustrated this by looking at IL13, a gene that encodes a cytokine secreted by T helper cells that in turn mediate allergic inflammation. Looking at the phenotypic associations for variants in IL13, 23andMe researchers found a clustering of associations around asthma, certain types of food and animal allergies as well as skin conditions like psoriasis. And indeed IL13 is a known target for several drugs that are currently in phase II and phase III clinical trials.
“At 23andMe, we are turning the drug discovery process on its head,” said Fah. “Traditionally a pharmaceutical company starts with a disease of interest and combs the disease pathways for drug targets. We start from a set of druggable molecules and use our genetic database to guide us to the disease indications. Because we scan across a wide spectrum of disease and traits, we can choose the most promising targets to work with.”
This is still early in this new drug discovery approach, he said.
“We feel incredibly blessed to be working with this powerful database, with our customers as partners,” Fah said.

Fah was not the only 23andMe scientist at American Society of Human Genetics this year. As we do each year, 23andMe had a strong showing, and our data was included in a number of presentations. This year we had two posters that were part of the conference.
– “Deep Learning and the prediction of human disease risk,” by Nicholas Furlotte, Babak Alipanahi and David Hinds
– “Improving haplotype phasing accuracy using many short IBD segments,” By Aaron Kleinman, Eric Durand and Cory McLean