The National Institutes of Health has funded a promising new study by scientists here at 23andMe that could help alleviate some of the existing disparities in genetics research between people of European and non-European ancestry.
Some estimates show that more than 90 percent of the research into the genetics underlying disease is on individuals of European descent alone. There are several reasons for this – historical, cultural, economic, and social. The lack of diversity has meant that non-Europeans have not benefited from the huge advances in genetic research. Still, by leveraging 23andMe’s large and ethnically mixed customer base, researchers here think they can change that.
“This could help reduce the genetics research disparities among groups in the United States and elsewhere,” said Kasia Bryc, the senior scientist and population geneticist at 23andMe who will lead the research. “The scientific impact of findings from this research may be significant.”
Instead of using traditional genome-wide association studies to identify genetic variants associated with disease, this approach, called mapping by admixture linkage disequilibrium, or admixture mapping, takes advantage of the history of population mixing in the United States. Although admixture mapping was first suggested decades ago, leveraging 23andMe’s database would take it to a larger scale. 23andMe has more than 1.2 million customers, 80 percent of whom have consented to participate in research.
Although most of our customers are of European ancestry, our customer base includes large numbers of African Americans and Latinos. 23andMe has also gotten very good at estimating ancestry along the genome, which is crucial for detecting associations with disease via admixture mapping.
Mapping by admixture linkage disequilibrium works by tapping into variants or sets of variants found more frequently 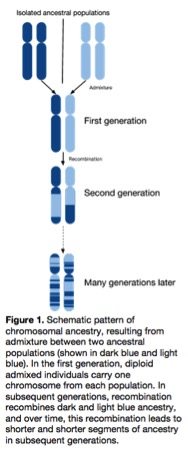 within one ancestral group. In admixed populations – such as African Americans and Latinos who have mixed European, African, and Native American ancestry – using this method may be a way to more easily identify disease-causing variants.
within one ancestral group. In admixed populations – such as African Americans and Latinos who have mixed European, African, and Native American ancestry – using this method may be a way to more easily identify disease-causing variants.
Looking at individuals with specific diseases and where there is “enrichment” for one ancestry should highlight genetic regions associated with that disease, or so researchers hope. This is because the disease-causing variants occur more frequently in segments inherited from ancestral populations with a higher frequency of disease-associated variants.
23andMe’s research will determine whether admixture mapping could become a powerful tool complementary to large-scale genome-wide association studies.
Our researchers believe this work could help uncover genetic variants associated with disease in both people of European and non-European ancestry. However, because much of the genetic variation in Europeans has already been discovered, we expect this study to be most valuable in providing insights into biological variation for non-Europeans.
Beyond what the study could do to alleviate some of the huge disparities in health research among non-Europeans, the work could also help point the way in other areas, including guiding the development of drug targets.



