We are all unique, and knowing our genotypes at specific places in our genomes may give each of us a better understanding as to why.
with an alphabet made up of just four letters – A, T, C and G.
Those four letters represent the nucleotide bases – adenine, thymine, cytosine and guanine – that are the building blocks of life. The sequence of A’s, T’s, C’s and G’s run in long stretches of code that instruct the way the human body is built and functions.
In 2003 after ten years and $3 billion, scientists decoded the first sequence of a human genome. Since then costs have dropped for sequencing a genome to as low as $1,000. While very inexpensive in comparison, it would still be prohibitive for most consumers to have their full genome sequenced.
So how is 23andMe able to provide genetic data to so many people at an affordable price in just a matter of weeks?
The secret lies in a process called “SNP genotyping.”
If you compared two unrelated individuals, you would find that their genetic sequences are about 99.9 percent the same. It is that miniscule variation in their genome that makes all the difference. It’s what makes us each unique.
That small variation is also the key to how 23andMe can efficiently and cost-effectively deliver its service. Scientists have identified the known variation among individuals, specifically locating in the human genome where a single letter differs from one person to the next.
These one-letter variations are called single nucleotide polymorphisms, or SNPs (pronounced “snips”), and they are associated with differences in everything from the color of our eyes, to our risks for certain health conditions, and information about the makeup of our ancestry.
Let’s look at an example of a SNP.
Say a certain percentage of people have a specific stretch of DNA code that reads ATGCCCGT, but everyone else has the sequence ATGCACGT along that same stretch.
The nucleotide bases come in pairs, so in this case the difference is between whether a person is CC or AC at this position.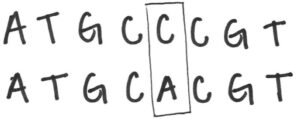
Since these two letters are known to vary among individuals, we would call this position in the sequence a SNP. The difference between two people at a single position – whether you are CC or AC in this case – is also referred to as the genotype for that SNP. Some of those differences may have no effect, some could be beneficial and some could be deleterious.
The benefit or harm of one version or the other can even change depending on one’s environmental surroundings. 23andMe provides individuals with information on hundreds of thousands of these SNPs.
To do that, 23andMe first has to extract DNA from saliva submitted by a customer. Once the DNA is extracted, it is amplified many times so that there is enough DNA to analyze. The amplified DNA is then cut into smaller pieces and washed over a microarray chip.
This genotyping chip contains thousands and thousands of tiny probes that are designed to detect specific bits of DNA. Each probe on the chip contains a bit of DNA that matches a genetic variant of interest. Remember the nucleotide bases always come in pairs, an A always pairs with T, while a G pairs with C. On the microarray chip, when a piece of DNA finds its complementary probe, it will stick to it.
These probes are triggered to glow in a way that indicates which version of each SNP is present in the sample. That fluorescent signal is read by a computer, and the data is then uploaded to the customer’s 23andMe account.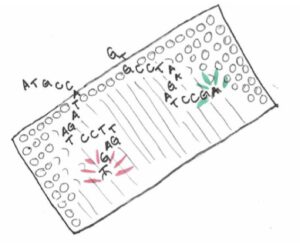
Our customers have the ability to view this raw data in a feature called Browse Raw Data.
Our latest genotyping chip, v4, is unlike any of our previous genotyping platforms. Previous genotyping platforms included standard probes included by Illumina, the manufacturer of the chip, as well as additional probes selected by 23andMe.
The v4 chip, consists of a completely custom panel of probes that were hand picked by our researchers. Although our current genotyping chip contains about half a million SNPs (whereas the entire human genome has around 10 million SNPs), we can learn a lot about the human genome simply by looking at these locations where individual genotypes are known to differ. Full genome sequencing provides information on all 3 billion markers in the entire human genome, however it is difficult to predict when full sequencing will be available at an affordable price point for the general public.



