
David Poznik
In the largest study of genetic variation within the human Y chromosome, an international team of scientists has traced back branches of the male line almost 200,000 years, revealing points in history of major population expansions that appear to correspond to eras of innovation and human migration.
David Poznik, a Computational Biologist and Population Geneticist here at 23andMe and the lead author of the paper, worked on the study while completing his graduate research in Stanford’s Program in Biomedical Informatics. Scientists from the Wellcome Trust Sanger Institute in the UK and from more than a dozen other institutions also contributed to the study.
Because the Y chromosome is passed in whole from fathers to sons through the generations, it offers a unique opportunity to reconstruct migrations that have taken millennia to manifest. By analyzing changes that have slowly accumulated over time, the 42 scientists were able to uncover a hidden history within the DNA sequences of 1,244 men representing 26 populations from around the world. The team used more than 60,000 Y-chromosome SNVs (positions at which at least one sequence differs from the others) to construct a tree that indicates how all the Y chromosomes are related to one another. Using the tree, they were able to follow modern male lines backward, ultimately tracing back almost 200,000 years to the most recent common paternal-line ancestor of almost all men currently living today.
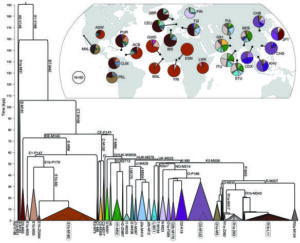
Tracing back in time paternal-lines from 26 different populations worldwide.
One notable feature of the Y-chromosome tree is that the different male lines naturally cluster into what are known as “paternal haplogroups” or “clades.” These groupings often correspond to different continental populations and can distinguish, for example, someone with Native American paternal ancestry from someone whose paternal line traces to Western Europe.
As a working group of the 1000 Genomes Project, the researchers had access to full sequences, which they compared to one another to measure distances between different groupings or clades. Since mutations steadily accumulate, these sequence distances correspond to temporal distances, with greater dissimilarities between sequences corresponding to longer separation times between their respective groups. Using published estimates of the Y-chromosome mutation rate, they estimated the timings of divergences between different haplogroups.
At certain points in history, particularly around 50,000 years ago and again approximately 15,000 years ago, several different lineages branched off from one another at the same time, marking major bursts in population growth. The times of these bursts overlap with human migrations into Eurasia and into the Americas, respectively. Between approximately 8,000 years ago and 4,000 years ago, there were other distinct periods of expansion in sub-Saharan Africa, South Asia, and Western Europe.
While the specific causes of those expansions are less clear, Dr. Chris Tyler-Smith, from the Sanger Institute believes that “the best explanation is that they may have resulted from advances in technology that could be controlled by small groups of men. Wheeled transport, metal working, and organized warfare are all candidate explanations that can now be investigated further.”
It will be interesting for scientists to use this DNA history in conjunction with archeological evidence to reconstruct the history of human migration across the globe.
It will be interesting for scientists to use this DNA history in conjunction with archeological evidence to reconstruct the history of human migration across the globe.



