By Jey McCreight, Ph.D., Research Communications Scientist
It takes a village to do research. Scientific discoveries from the 23andMe Research Team are made possible both by our three million research participants and by our collaborators. We sat down to talk with one of those collaborators, Louis Muglia, M.D., Ph.D., Professor of Pediatrics at the University of Cincinnati, Vice Chair for Research, Director of Human Genetics, and co-director of the Perinatal Institute at Cincinnati Children’s Hospital Medical Center. Dr. Muglia lead the recent collaborative study on preterm birth that was published in the New England Journal of Medicine.

Jey McCreight: What motivated you first to start studying gestational time and preterm births?
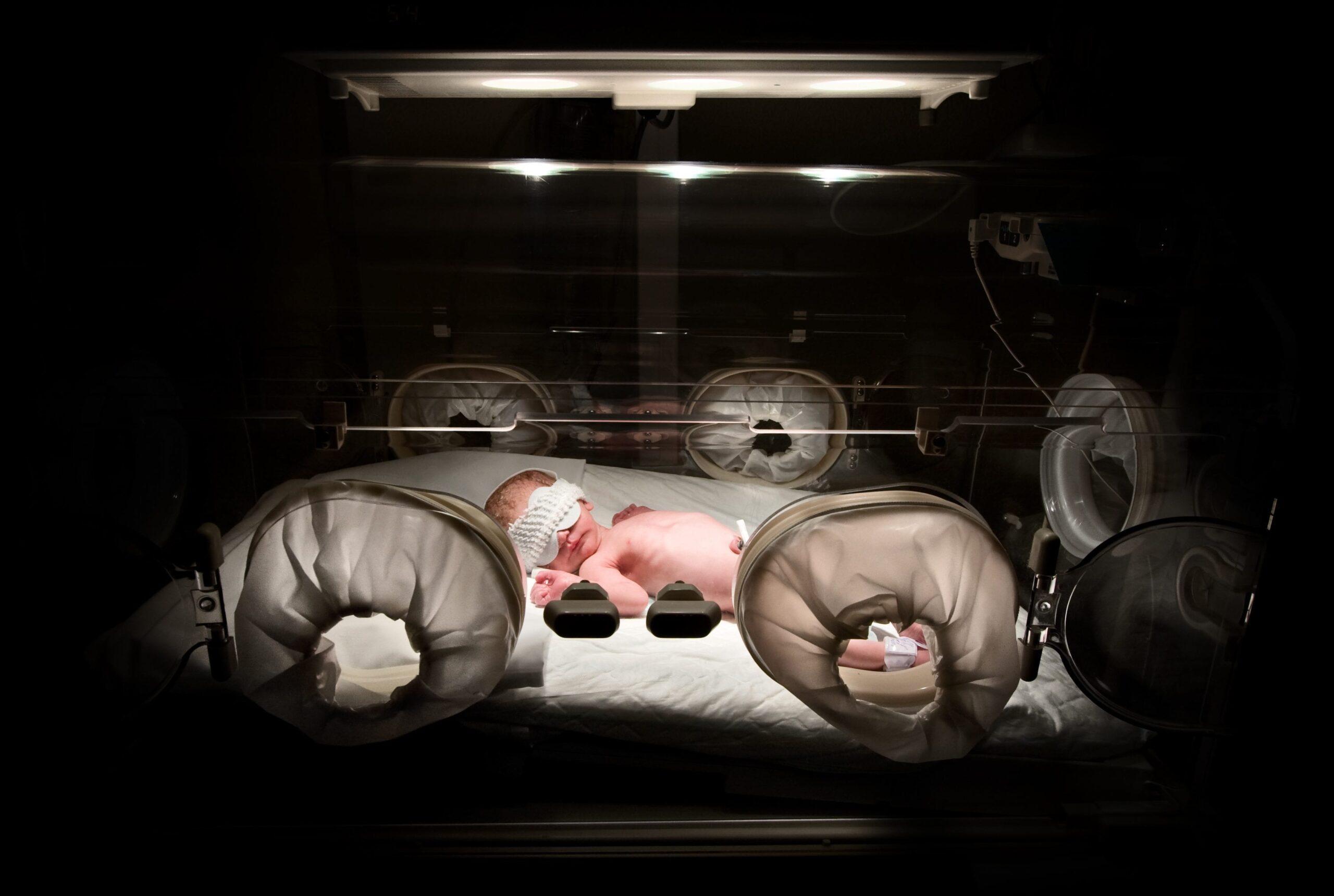
Louis Muglia: Well, I’m trained as a pediatrician initially. When I was a resident in Boston, we spent a lot of time in neonatal intensive care units, and that really made a lasting impression on me. I mean, you see these rooms full of tiny babies that are born at 23, 24, 25, or 30 weeks gestation that are suffering huge medical problems, who are many times either dying or devastated for life. The only reason they have those problems is that they were born at the wrong time. And as you probably know, preterm birth has been the leading cause of infant mortality for many, many years. But it’s recently become the leading cause of all child under five mortality because we’ve made more progress in other areas.
And we don’t understand why birth timing occurs when it does in women. We don’t know the mechanisms that get us there. To me, as an endocrinologist as well – I did pediatrics and pediatric endocrinology – pregnancy is a great endocrine physiology system. Hormones are changing like at no other time in a person’s life except when they go through puberty. It’s just a fascinating way to think about getting your handle on what’s causing those regulatory changes, because in my mind, doing genetics and genomics is really the only way you’re going to get our foot in the door in a non-biased fashion to understand molecular pathways. Because you’re not going to do anything that disrupts a pregnancy. In my opinion, it’s the single most important problem we have left to solve in maternal and child health.
JM: So what made you first interested in collaborating with 23andMe?
LM: I say it was the potential to utilize existing data that could drive discovery ahead in a way that would take extraordinary resources and an inordinate amount of time to do without them. We didn’t know what data was available from 23andMe related to pregnancy … but I figured why not? I mean, they’ve got millions of research participants, and even if there was some small fraction of them who had answered a question relating to pregnancy outcomes, it would be a shame not to be mining that information and using it to shape our thinking.
JM: Did you have any initial concerns about working with a consumer genetics company or using self reported phenotypes when you were getting started?
LM: Absolutely. The concern I had was that we would just get noise — you know, the self-reported data, especially around the length of pregnancy … And the other thing that we were specifically interested in was understanding why women went into labor spontaneously. We weren’t as interested in women who delivered via C-section because they had pre-clampsia, or the baby wasn’t growing well, or there were other medical complications in the pregnancy. And I thought those two factors with self-reported gestation length were just going to make the information a little bit too heterogeneous for us to be able to get robust associations.
I wasn’t so worried about false positives, though. Because, something that emerged from it, we were going to validate in our carefully phenotyped replication cohorts anyway. So we knew we had 8,000 moms who we could look at who were well phenotyped to determine whether the hits we were getting with the 23andMe data and self-reported data, when we weren’t exactly sure on the mechanism of pregnancy outcome, was actually giving us the information we thought it was.
People had told me that large numbers of samples kind of trump the concerns with noisy data. I didn’t believe it. I absolutely believe it now. I’m a firm believer that big numbers of samples make all the difference.
JM: We’ve converted you?
LM: Heh, well, truly.
JM: What’s your opinion now after collaborating with 23andMe? Did anything change as you actually went through the process with us?
LM: Yes, this has shaped the way I think about how we should be collecting data that’s available to us every day. Anything that we’re doing on a large-scale population level with self-reported data, we should be capturing the information from these really critically important medical issues and not worrying so much about whether the reporting quality is perfect, but just getting the big numbers of subjects. So things like the All of Us Initiative that’s coming out from the NIH, the Million Veteran Program, all of these things — we need to make them bigger and ask the right questions associated with them, and hope we get enough people answering the questions, and not worry so much about whether every answer is going to be perfect.
And you know, it makes me want to explore more opportunities with 23andMe. Our first pass at this was looking at the mom’s genome because we had the most evidence that there were findings there. I’d love to go back and develop a plan — we’re just working on this right now — to look at the fetal genome. There’s not a question on the survey right now that lets us do that. They don’t ask the individual who’s getting their DNA tested at 23andMe “How many weeks gestation were you born at?” They ask the mom “How many weeks was the gestation of your first pregnancy?” That gave us the mom’s genome information related to pregnancy. So I’d love to go back and build in a question into a survey that actually asks that information.
JM: How has the rapidly increasing availability of large datasets like ours changed your research over the years compared to what you were doing earlier?
LM: Well, it’s really made humans our animal of choice to do investigation on. The benefit we’ve had in the past – and this is my background – is I was a mouse molecular geneticist. I did gene targeting in mice, taking specific pathways, asking how they affected physiology, and then trying to take that and extrapolating it to human biology. And the rate limiting factor was, when you try to extrapolate it, it would never line up right. You just couldn’t. The physiology had changed too much between typical laboratory animal species like mice or sheep or rats, and women.
So we eventually had to have the ability to study human pregnancy. It’s not until really big datasets like this became available that we’re really able to get into pathways that we can ask how common variants affect these outcomes. And for pregnancy outcomes, you might guess that they’re going to undergo the most evolutionary selective pressure of virtually any trait, right? I mean, that’s reproductive fitness that’s being selected for. Any common variant that exerts a big detrimental effect during pregnancy is not going to be around for very long. So you’re looking at things that are going to have relatively small effects, and probably have beneficial effects in some other way that protect you from some other human malady. So I think you need big data sets to reveal those small effect sizes, and I think with the 23andMe collaboration that was proven wonderfully for preterm birth.
JM: You were bringing up natural selection, which totally makes sense in regards to this. I noticed that you had previous studies where you used comparative genomics to identify human genes involved in birth timing that were rapidly evolving with respect to other primates.
LM: Right.
JM: But those genes didn’t seem to be the same ones that you had identified in your recent study, and I was wondering why you thought that might be?
LM: I think we really didn’t achieve genome-wide level significance in those initial studies. So we did a dimensionality reduction by just selecting out the most rapidly evolving genes. And I think those findings are incredibly interesting and still quite useful in terms of thinking in context, but we did that study in a relatively limited population from Finland. I think that initially heavily targeted our data. The larger population that we did affected a much more diverse European ancestry population than we originally would get in Finland. I think when you start doing these big data sets, the most robust sequences emerge. And it turns out the ones that are emerging are probably not the ones that are under the most rapid selective change in humans. I think some of them are, but many of them are part of a toolbox that is conserved and functioning in many ways. The genes we got like the WNT family that came up in the paper, they’re involved in many aspects of development that are very hard to have rapidly evolve because you’re going to affect too many fundamental developmental pathways.
JM: You mentioned that some of your previous studies were done in Finnish populations, and I know this particular study was still limited to women of European ancestry. Are you planning on following up with any non-European populations? Because I know from some of your previous research that women of African American ancestry are four times more likely to have recurrent preterm births.
LM: Yes. What we wanted to do initially for gene discovery was evaluate cohorts that had the least environmental risk for preterm birth — so, you know, had good maternal care, were not under socially deprived circumstances. We know that that’s not the most at-risk population, but we think that it probably allowed us to more sensitively detect the genes associated with the outcomes. And now what we’d like to do is go back (and look at) other ethnicities and begin to ask “Are those same genes associated?” We’re starting to do that in existing cohorts of women that have been collected, but we’re also embarking on a big study that’s being supported by the Gates Foundation to do genome wide association in about 10,000 African ancestry women and South East Asian ancestry women.
JM: Fantastic. Well that’s exciting future research to look out for.
LM: Yeah, you know, super important. My guess is we’ll get many of the things that replicate in African ancestry populations. I don’t think that I have any doubt that they won’t, but I think we’ll get new things too that reflect different environmental pressures in places with higher susceptibility and higher rates.
JM: One thing that came out of this study was the gene EEFSEC, which involves using selenium. You were proposing that this potentially could be a test for identifying women at risk for preterm births. Do you have any research plans to follow up with that in the future?
LM: Yes, and this is part of the same study that the Gates Foundation is supporting. So we identified the gene you mentioned, EEFSEC, which is involved in generating selenoproteins. And again, this is why you do genome-wide associations: This was not anywhere on my radar screen, thinking about selenoproteins involved in pre-term birth, but they make a lot of sense. They’re antioxidant, they’re anti-inflammatory, they probably protect from premature aging of gestational tissues. We don’t know exactly how they’re working, but in this low environmental risk population, we identified this gene involved with selenoprotein metabolism as being associated with preterm birth risk. That sort of raised the notion of whether dietary deficiency in other populations that are potentially at risk for inadequate intake of selenium, which is an essential micronutrient, could that be driving their risk for preterm birth in addition to the genetic predisposition. So right now we’re doing a big epidemiological study in low- and middle-income countries where we’re looking at the relationship of maternal selenium levels halfway through pregnancy with the risk for preterm birth at the end of the pregnancy.
JM: You were saying earlier that your previous research used mouse models. Do you plan on returning to mouse models to further study some of the genes implicated in this study?
LM: Yes, for sure. The thing that we did in the mouse was we started out picking genes we thought might be important and then seeing if they were important for the mouse — and sometimes the answer was yes, sometimes the answer was no — and then going to human. What we’re doing now is we’re taking the SNPs we’ve identified in humans, and we’re going to do two things: We’re going to in vitro models to look to see if they alter regulation of the associated genes, and what that does to network expression of RNAs in cells, but we’re also taking those and engineering the variants into mice using CRISPR Cas9. Because many of these, even though they’re in non-coding regions, are highly conserved between humans and mice, and it will give us an inroad into what it’s doing in pregnancy duration.
We just did this for our first, the WNT4 allele that we identified that we worked up in the New England Journal paper. We think we identified that estrogen receptor binding site as being the causative mechanism for the change that we were seeing. That region is 98 percent conserved in mice over a 50 base pair region. We engineered the one point variant in, and we just have our founder lines to be able to do that analysis now. So we’re hoping that that will be an informative approach moving forward. For some other experiments that we’re doing, we’re doing genome editing in non-human primates, so we may go to that as well.
JM: Exciting. The CRISPR revolution has even touched your research, I see.
LM: Absolutely, we do a lot of CRISPR Cas9 stuff.
JM: Are there any other research projects that are on the horizon that we haven’t touched on yet that you’d like to share with us?
LM: I think one of the things that is really interesting to think about is classically we’ve always thought of the mom’s genome as sort of being one unit for analysis and the baby’s genome being another unit for analysis. But I’m really hoping to get to this notion of looking at what I would call a combined genome of pregnancy — a metagenome of pregnancy — where you have both maternal and fetal genomes interacting in a way that mutually shapes pregnancy outcomes. So mom’s genes program maternal environment which then have effects on fetal health, but my guess is that there are fetal genes that shape mom’s pregnancy environment that affect mom’s health. I think looking at these bidirectional signaling pathways between the mom and the baby are something we’d really like to do to really fully understand how you optimize health in human pregnancy.
JM: That would be really exciting to look at — I hope that study works out, and all the future ones.
LM: Yes, we’re really excited about doing that. What you need for those studies are maternal-infant dyads, largely. And probably the way we’re collecting information with 23andMe would make it a challenge to be able to do that — maybe not impossible, but still a challenge.
JM: I know we have trios (parents and a biological child) in our database, so hopefully there’s some data in there for you.
LM: Yeah, that’d be great! I think the other thing is integrating; now we have a solid foundation for a handful of genes that are involved in human pregnancy. We can begin to now make sense of many of the gene expression and other omics datasets that have been collected around pregnancy to really nucleate on the core set of genes we’ve identified. Because that’s actually the most solid data that we have related to essential elements of pregnancy timing for humans.
JM: That’s great. Is there anything else you’d like to share that we haven’t covered yet?
LM: No, you know, I just thank 23andMe for all that they’re doing to collect this kind of information and make it available to the research community. It’s just such a fantastic resource, it really is. I think the more we can use this as a model to gather information that can really change health, by what we’re doing every day anyway, I think we’re going to accelerate our progress enormously.
JM: Well thank you, that’s our goal too, and we love collaborating with researchers and advancing science.
LM: You guys have been terrific to work with.