By Samantha Ancona Esselmann, Ph.D., 23andMe Product Scientist
Are you at increased likelihood for high blood pressure, gout, or something else? Here’s how 23andMe determined your result…
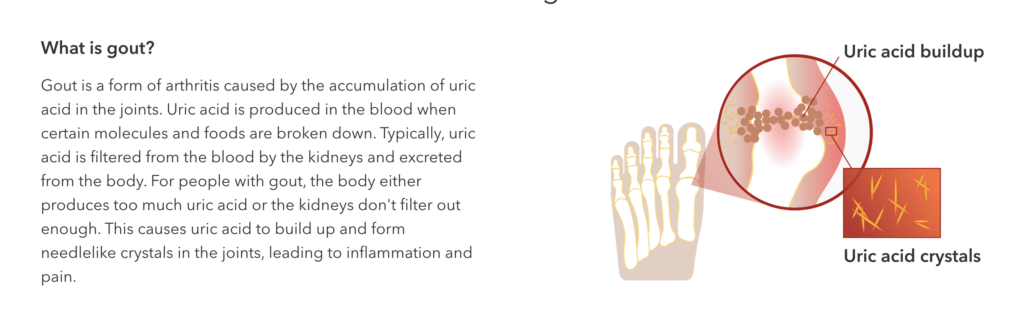
A few weeks after California entered its first pandemic lockdown, my mother called to tell me she’d felt a sudden, stabbing pain in her foot. It hurt so much that she thought she’d broken a bone, and a frantic trip to urgent care revealed she was likely experiencing symptoms of gout.
My mother was shocked by the diagnosis. She’d read enough historical novels to know that gout was once called “the rich man’s disease” and “the disease of kings,” thanks to its association with alcohol and rich foods like red meat. How could she—a woman who lives by the motto “moderation in all things”—have a condition long associated with excess and intemperance?
It made me wonder if gout has anything to do with genetics.
In October 2020, just a few months after my mother’s diagnosis, 23andMe released a new report for 23andMe+ members, powered by consented 23andMe research participants, that identified new links between genetics and gout. My report told me that I had an “increased likelihood” of developing gout, and that an estimated 19% of female research participants with genetic results and ancestry like mine are expected to develop gout by their 70s.
Given my mother’s diagnosis, I wasn’t surprised by my increased likelihood, but I was surprised to learn that my result was generated using over 20,000 genetic markers — also known as DNA variants. How could the combined effect of so many factors be analyzed all at once?
So, I “sat down” (virtually) with other 23andMe scientists who walked me through how 23andMe creates these cutting-edge predictions. It’s relevant not just for gout, but also for understanding how 23andMe creates its Type 2 Diabetes report, or our recent report on eczema (atopic dermatitis). In each case the reports analyze hundreds, thousands, or tens of thousands of different DNA variants all across the genome to calculate the likelihood of developing a condition.
Here’s what I learned
Making 23andMe’s polygenic score for gout
1. First, we study DNA variants associated with gout.
23andMe scientists compared the DNA variants of many 23andMe research participants who said they have gout to the variants of many participants who said they don’t have gout. During this process, the scientists discovered thousands of variants across many different genes that are more common among people with gout, which means those variants are considered to be “associated” with gout.

This process is called a “Genome-wide Association Study,” or simply, a “GWAS” (pronounced Jee-Wahss). DNA variants found in a GWAS can provide clues about the biological mechanisms underlying a condition. Those variants can also help scientists estimate the likelihood of someone developing a condition in the future.
What else does a GWAS tell you?
- DNA variants identified in the GWAS can be associated with either an increased or a decreased likelihood of developing the condition.
- Some of the variants found in a GWAS can be very strongly associated with the condition, while others may be only weakly associated. The strength of the variant’s association with a condition is also called the variant’s “effect size.”
- A weak association with a high statistical confidence usually means the variant has a small, but real effect. A low-confidence, weak association could mean that there were simply not enough people in the study to allow researchers to confidently identify variants with small effect sizes.
- Just because a DNA variant is statistically associated with a condition doesn’t necessarily mean that it causes the condition. That’s because variants that are physically close to each other in your genome tend to be inherited together, which makes it difficult to figure out which of the linked variants actually causes increased likelihood. But, that’s okay! For the purposes of predicting a condition, researchers don’t need to know which variants are biologically involved, as long as they find a selection of variants in the same genetic “neighborhood” as the ones that cause the condition.

2. Next, a computer trains itself to predict gout.
Now we have a list of DNA variants associated with an increased likelihood of gout in a group of folks who already know they have gout, so we can try to use that information to predict whether other people are likely to develop gout.
How do we do this?
Various combinations of DNA variants associated with gout are fed into a computer algorithm.

In this step, called “training,” the computer’s goal is to use the DNA variants found in the genome wide association study as a reference to help it learn how to identify patterns in the data and to predict gout correctly in a group of people whose history of gout is already known.
At this stage, the trick is that the computer technically knows the “truth” about who has gout and who doesn’t have gout. This way, it can quickly check whether or not it’s getting better at predicting gout.

But, understanding the combined impact of thousands of variants to predict gout is no simple task. Just think about how complicated it can be to guess the price of a home: There are lots of factors that can impact a home’s price, from square footage and number of bedrooms to neighborhood comps. In the real world, these variables are not independent, and they can impact each other in really complex ways, for example: The relative importance of square footage might be lower in a walkable city center than it is in the suburbs.
On its first try, the computer probably won’t be very good at predicting anyone’s likelihood of developing gout. After all, how good would you be at guessing the price of a home based on each of its features if you’d only ever seen one listing?
So, the computer goes back to the drawing board, rapidly adjusting and tuning the impact, or “weight,” of thousands of DNA variants until it finds the best set of parameters to predict gout.

For any person, the computer will add up the combined, adjusted weight of all the variants and assign a likelihood score, called a “polygenic score” (polygenic means “many genes”).
During this “training” stage, if someone had previously told us they had gout, they would be more likely to receive a high score. If someone told us they did not have gout, they would be more likely to receive a low score.

When the computer thinks it has found the best possible combination of parameters for the “training” group, we’re ready to test how well it performs.
3. Then, we test how well the computer predicts gout.
Before the computer graduates from training, we test its performance in a new group of people, some of whom have told us they have gout and some of whom do not have gout.
Why do we do this? We perform this step because the computer sometimes overestimates its ability to predict gout in the training step, and there’s a chance that the group of people we selected for training the computer is biased. For example, what if you trained a computer to predict the sale price of a house, but you only trained it on houses with one bedroom? When you later ask the computer to predict the price of a four-bedroom house, it could fail because it hasn’t encountered large houses before.
At this testing stage, we also make sure to adjust and validate the computer model for people with ancestry from different parts of the world.
4. Your likelihood of developing gout is reported to you.
Now, we have a computer algorithm that can predict likelihood for gout based on over 20,000 DNA variants. At this point, the trained computer algorithm is ready to estimate your likelihood of developing gout by analyzing your unique combination of DNA variants.
We send your DNA variants through this trained computer algorithm, and it computes your likelihood of developing gout as a polygenic score.

If the odds associated with your score are at least 1.5 times higher than the average odds, your likelihood is reported to you as “increased.” If your odds are less than 1.5 times higher than average, then your likelihood is reported to you as “typical.”
Your personalized polygenic score is then translated into a lifetime chance of developing gout for people with genetics, self-reported sex, and ancestry like yours.
Gout fast facts
- The T-Rex named “Sue,” discovered in South Dakota in the 90s, may have suffered from gout.
- Galen, a 2nd century C.E. Greek physician and philosopher in the Roman Empire, recognized gout as something that could be inherited.
- The word “gout” comes from the Latin word gutta, meaning “a drop,” a reference to the historical belief that too much of one of the four bodily humors would “drop” into a joint, causing pain. The word “gutter” also comes from gutta.
- Gout may have precipitated the U.S. Revolutionary War. Suffering from symptoms of gout, British parliamentarian William Pitt the Elder was absent from British parliament when new forms of taxation were being imposed on British colonies in the Americas. Had Pitt (who was famously opposed to taxing colonists without giving them parliamentary representation) been present in parliament during these sessions, he may have been able to block the inflammatory legislation, possibly preventing — or at least delaying — the Revolutionary War.
Putting it all together: what does it really mean for me?
23andMe tells me that an estimated 19% of female research participants with ancestry and genetic results like mine develop gout by their 70s, which puts me squarely in an “increased likelihood” category.
Does this mean that if there were five carbon copies of me, one of them would definitely develop gout in her life?
Not necessarily. Instead, it means that one in five women with ancestry and genetics like mine will develop gout in their lives.
My colleagues urged me to remember that every female of European descent whose data contributed to this prediction has her own unique lifestyle, family health history, and genetic ancestry. Some drink excessively or eat red meat every day. Some are underweight. Some are obese. Some have recent ancestors from Ireland, while others have recent ancestors from Greece.
And all of that variation gets compacted — flattened into a single output. That means it’s possible that my family history, lifestyle, and ancestry could affect my chances of developing gout in ways that 23andMe’s prediction — based mostly on genetics — simply cannot account for.
In the future, scientists at 23andMe would love to develop the ability to account for variables like environment, lifestyle, and family history in addition to DNA.
If my ancestry weren’t European, would this result be less accurate?
Okay, real talk. Historically, it has been easier for academic groups and companies like 23andMe to provide more accurate genetic likelihood estimates to people of European descent. Why? Because there have been far more genetic studies on individuals with European ancestry to date. And, generally speaking, more data means better computer models.
But, at 23andMe we want to release reports that also work well for people of non-European ancestry. So, how are we working to improve results for customers with non-European ancestry?
- While it’s true that the majority of the 23andMe database is composed of people of European descent, 23andMe has the most Latino/Hispanic and African American research participants of any genetic database in the world.
- This means we can adjust and test our computer models on people from many backgrounds to make sure they also work well for non-Europeans.
- As more people with non-European ancestry choose to participate in 23andMe research, our ability to estimate genetic likelihood for many traits and health conditions will improve for people with non-European ancestry.
What’s the takeaway?
Simply this: compared to most other female 23andMe research participants with European ancestry, I’m somewhat more likely to develop gout in my life because of my genetics.
The bright side is there’s still an 81% chance that I won’t develop gout in my life, and I may be able to reduce my chances even more through lifestyle modifications. What can I do?

Limit my consumption of red meat? Easy!
Limit my alcohol consumption? Working on it!
Aim for low-fat or nonfat dairy? I’ll try, but I live with a German, so this could be tricky…
Maintain a healthy weight? It comes and goes…
And, if you were wondering…since her diagnosis, my mother has been more careful to drink lots of water and to limit her alcohol consumption.
She hasn’t had another gout flare-up since.
Read more from 23andMe about polygenic modeling in our blog:
- 23andMe Researchers Develop A Better Way to Estimate Skin Cancer Risk
- A Conversation with Nilanjan Chatterjee
Thanks to other members of the 23andMe product team for their insights and feedback, in particular Peter Chisnell, Becca Krock., James Ashenhurst., and Alisa Lehman.