Mexican flu virus genetics has been much in news lately; how about a look at Mexican human genetics for a change?
A new article from researchers at Mexico’s National Institute for Genomic Medicine (INMEGEN) examines genetic diversity across the nation and argues that, in order to conduct studies of common genetic diseases efficiently, a Mexico-specific genetic reference database should be built.
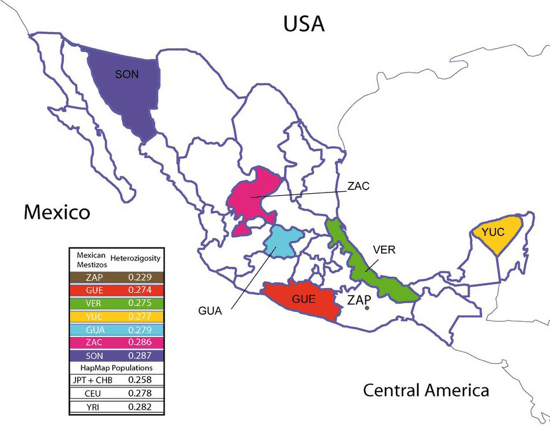
Map of Mexico with sampled-from states highlighted.
Recent studies of genetic diversity among Europeans (blogged about here ) show that DNA is a surprisingly good predictor of where a person lives; people from the same country tend to be more similar to one another than to those from other parts of the continent. This latest study, which was published earlier this week in the Proceedings of the National Academy of Sciences, shows a similar pattern in Mexico: Mexican Mestizos (people of mixed European and Native American ancestry) from the same state tend to group together genetically, and the groups themselves fall along a genetic continuum that corresponds roughly to their latitude. You can see for yourself in this plot from the paper (below), which we’ve modified a bit for clarity. This is the same kind of plot used in 23andMe’s Global Similarity: Advanced feature – each point in the plot represents a person. The closer two points appear in the plot, the closer those two individuals are to each other genetically. The 300 Mexican Mestizos fall into a line stretching from a group of Europeans at the upper right to a group of Amerindians1 at the lower left.
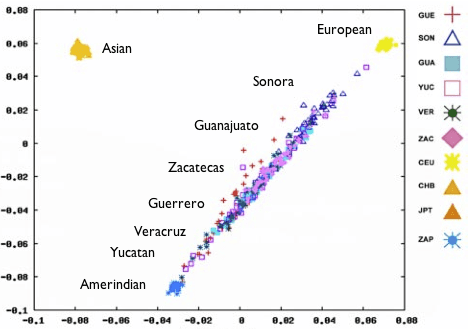
PCA map of Mexican genotypes.
The people from Sonora, the northernmost state, appear closest to the European cluster, and the people from the southern states Guerrero, Veracruz, and Yucatan appear closest to the Amerind cluster. This makes you wonder whether this pattern might correlate with the proportion of European ancestry. The researchers wondered that too, so they investigated further by analyzing their dataset with a computer program that estimates the proportion of ancestry a person’s DNA derives from each of several reference populations. When they set the program loose, they found that the six states did vary widely in proportion of European ancestry, from an average of 65% in Sonora (fifth column from the left) to an average of 35% in Guerrero (second from the right):
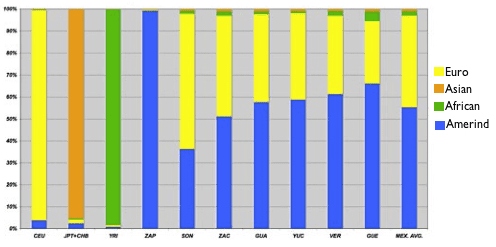
Admixture proportion estimates for Mexican and HapMap samples.
The authors note that this pattern makes sense since Amerindian population density declines as you head north. Also, you might note that there’s a green sliver of African ancestry in each of the Mestizo populations, which approaches 5% in the southern states of Veracruz and Guerrero. This also meshes well with the historical record, since it’s through these coastal states, among others, that African slaves entered Mexico during the Spanish colonization.
These analyses tell us much about Mexico’s history – could the same dataset serve to improve Mexico’s future? This paper also examines the prospects for conducting medical genetic studies in Mexico.
The most popular study design nowadays, and the one that the researchers focus on, is the genome-wide association study, or GWAS, also called a case-control study. GWASes are the basis for essentially all the studies discussed in the Blog, and the majority of the findings underlying 23andMe’s Health and Traits reports, and have recently been the subject of intense debate in the genetics community.
The core idea of a GWAS is to look for genetic markers (these days, usually common single-letter DNA variations known as SNPs) in a population that are at very different frequencies in a group of people with a particular disease, say type 2 diabetes, then in a group of people without that disease. In order to do that, you first have to identify a comprehensive set of the common genetic variations within a population, and then create a DNA array (commonly called a SNP chip) to probe all those variable locations in a large number of people with and without the condition you’re studying.
There’s the rub. In recent years, a project known as HapMap has created catalogs of common variations in European, African and East Asian populations, and chips have been produced based on it2. But Mexico’s population is a mixture of two of those (European and African) and another population (Amerindian) that are related, but not identical to, the third. There is no Mexican SNP chip.
That’s why the authors of this paper are suggesting a project to characterize common genetic variation within the Mexican population itself. They estimated that a catalog of common genetic variation using any two of the Mestizo groups they analyzed would capture enough variation to fuel quality GWAS studies, and would require fewer markers to do so than the alternative, which would be to use all the common markers from HapMap itself. This would substantially lower the cost of genotyping, they argue, and the reduced cost of using a platform based on the Mexico-specific catalog would allow researchers to genotype many more people for their GWAS studies for the same number of pesos. Since the sample size is often the limiting factor in the ability of the GWAS design to find disease genes, this could improve their ability to find the genetic causes of inherited disease.
Thanks to 23andMe Founding R&D Architect Brian Naughton for his assistance in the preparation of this blog post.
Notes
- Amerindian, sometimes just Amerind, is short for “American Indian”, and it denotes a descendant of the indigenous peoples of the Americas; anthropologist-types use the word to avoid confusion with the Indians that live in South Asia.
- Because full-genome sequencing is still a few years from being affordable, researchers cannot look at every single one of the 3 billion nucleotides in the genome to find the one (or combinations of more than one) that are directly linked to a particular condition. For now, they must make do with the half-million to million SNPs that the current crop of SNP-genotyping chips allow. At first blush, this sounds like a fool’s errand. How can you possibly say anything about 3 billion DNA nucleotides with a collection of just a million markers? What if the marker you have on your SNP chip is near to but not actually the disease-causing SNP? Won’t this be like ships passing in the night? In just the last decade or so, geneticists have learned that the human genome has a very peculiar property: the 22 numbered chromosomes, or autosomes, and the X, tend to do the bulk of their recombining in a very small fraction of the spans of those chromosomes. These highly-recombining locations are termed hotspots; it’s kind of like chromosomes are trains, with hotspots as the links between boxcars (although unlike boxcars, which are all the same length, there’s considerable variation in the distance between hotspots). This state of affairs is good news for GWASes, because it means that these boxcars – big chunks (mean length ~200,000 DNA base pairs) of chromosome that tend to be passed from parent to child as a unit – aid the task of marker selection greatly. If you sample a bunch of people, as these researchers have done in Mexico, then you can build up a catalog of the specific chromosome chunks that occur. The idea is that when a disease-causing allele occurs, it can’t help but sit on one of these chunks, so the problem of building a GWAS chip is transformed into choosing markers that reliably distinguish between the chunks. It’s like being able to assign one inspector to each boxcar, rather than each crate inside it. All you need to do, then, is to look at SNP that’s diagnostic for the chromosome chunk that your disease-causing SNP is sitting on, instead of having to find the causal SNP itself. The technical word for these chunks or boxcars is haplotype3, and such catalogs when built are called haplotype maps; it’s actually what was meant by the “medical genetic reference database” in the title of this post. It’s for the same purpose that the International HapMap Project, which has built reference haplotype maps for African, Asian, and European populations, was conceived, and it’s the origin of the name HapMap.
- A haplotype is short for haploid genotype. Haploid means that you’re concerned with one chromosome, so haplotype means a contiguous segment from a single chromosome. Diploid means two (paired) chromosomes; humans are a diploid organism because our chromosomes come in pairs. The fun doesn’t stop there. Some organisms are tolerant of higher ploidy, so there are tetraploid, hexaploid, and even octoploid species. For example wheat is hexaploid, so it has six copies of each chromosome.