A recent study in The Lancet reported that variation on the Y chromosome — haplogroup I, tagged by the SNP rs2032597 — is associated with coronary artery disease (CAD) in British men, with an odds ratio of 1.56. This finding stands out for several reasons. For one thing, the effect is substantially larger than any other reported genetic associations with CAD. Furthermore, it is rare to hear about functional effects of Y chromosome variation — at least partly because the Y chromosome is small and contains very few genes. The finding also invites special attention for some of the same reasons that make the Y chromosome interesting for population genetics. Here, we discuss the special properties of the Y chromosome and their implications for association studies, our concerns about the evidence the authors use to support their finding, and observations from the 23andMe database for haplogroup I and heart disease.
Why the Y is problematic
Most of the Y chromosome is not subject to recombination (the shuffling of maternal and paternal DNA that creates the unique genetic material passed down through sperm and egg) and is inherited intact from father to son. A consequence of this is that genetic variation on the Y chromosome is strongly associated with genetic ancestry. That doesn’t necessarily mean that genetic variation on the Y can’t also be functional, but it complicates functional analyses because it can be especially difficult to distinguish between ancestry effects and effects that are directly due to a particular Y chromosome variant. The lack of recombination also means that variants that are very far apart on the Y chromosome can still be strongly correlated, so it is also very difficult to use an association on the Y chromosome to identify a particular region as functionally important: the causal variant could be almost anywhere on the chromosome.
How strong is the evidence?
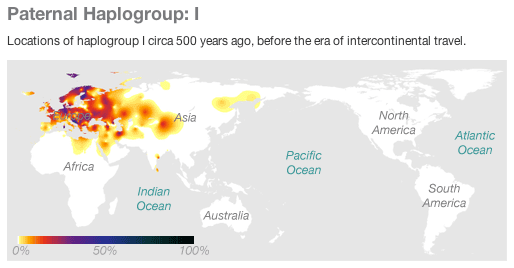
In The Lancet study, the authors carefully checked for alternative explanations for the association between haplogroup I and CAD. They looked for differences in conventional cardiovascular disease risk factors — both clinical biomarkers like blood pressure, BMI, and HDL, and lifestyle factors like smoking, alcohol consumption, and education — between haplogroup I and other haplogroups. None of these explained the association between haplogroup I and coronary artery disease. The authors also showed that the presence of haplogroup I did not correlate with the most significant dimensions of genetic ancestry within the study participants. This is a little surprising because haplogroup I is differentially distributed across Europe on a larger scale: it has a higher prevalence in Scandinavia and a lower prevalence in Southern Europe. Of course, it is possible that this gradient is not present in the smaller population of British men.
The trouble with candidate gene studies Candidate gene association studies (studies looking at one or a few genes as opposed to studies looking at variation across the entire genome) have a long and somewhat sordid history, largely because we tend to misjudge our ability to choose hypotheses that have a good likelihood of being true. If we choose a SNP at random and hypothesize that it might be associated with CAD, it would probably be better to use the genome-wide threshold for statistical significance (p-value < 5 x 10-8) to evaluate the results, rather than, say, P < 0.05 based on the fact that we have done just a single statistical test. If we’ve used a lot of prior knowledge to choose this one SNP, then maybe we could argue for using a more lenient standard. But it is very hard to decide how much prior knowledge is “worth” in this context.
What about the statistical evidence in favor of the haplogroup I association? In the target population of British men, more than 90% of individuals have either haplogroup I or another paternal haplogroup, R1b1b2. One could thus argue that the authors are effectively performing just a single statistical test comparing these two groups. On that basis, their statistical results (p-values of 0.004 in their initial study set, 0.012 in their replication set, and 0.0002 overall), seem convincing. However, it could be argued that the odds were stacked against their initial hypothesis that Y chromosome variation might be associated with CAD. The reported result would not meet criteria for significance in a genome wide association study (typically, p-values < 5 x 10-8), so a lot depends on the credibility of this initial hypothesis.
The authors suggest that their hypothesis was motivated by the observation that CAD is more common in men than women but this fact is not convincing by itself. Physical differences between men and women are largely determined by the presence or absence of the Y chromosome, but that does not necessarily mean that specific variations on the Y chromosome affect traits or risk for disease among men. (It is much more likely that variations in many other parts of the genome that are functionally involved in those traits account for those differences.) The authors also cite work on animal models, as well as a few studies in humans suggesting that Y chromosome variants may affect cardiovascular phenotypes. However, the authors did not test the specific variants or phenotypes implicated in those studies.
Findings in the 23andMe community
We investigated whether the tag SNP for haplogroup I — the C version of rs2032597 — was associated with cardiovascular phenotypes in men with European ancestry from the 23andMe community using four traits: self-reported coronary artery disease, heart attack, high blood pressure, and high cholesterol. The following table shows results of association tests for these phenotypes. We see no significant association between rs2032597 and any cardiovascular phenotype, and can easily exclude values as large as the odds ratio of 1.56 reported by the authors of the Lancet study.
Table 1. Association results for rs2032597 and heart disease phenotypes
| # cases | # controls | p-value | Effect | 95% CI | |
| Coronary artery disease | 1014 | 20120 | 0.92 | 1.009 | [0.845,1.206] |
| Heart attack | 486 | 22498 | 0.31 | 0.881 | [0.687,1.130] |
| High blood pressure | 5230 | 15797 | 0.34 | 0.958 | [0.878,1.046] |
| High cholesterol | 7970 | 15030 | 0.84 | 1.008 | [0.932,1.090] |
In contrast, we replicate the association for rs4977574, which is the variant (located on chromosome 9) most strongly associated with CAD in a recent large-scale meta-analysis. In this case, results from the 23andMe database match the reported odds ratio of 1.3, suggesting that self-reported data from 23andMe members is sufficient for detecting true associations.
Table 2. Association between rs4977574 and heart disease phenotypes
| # cases | # controls | p-value | Effect | 95% CI | |
| Coronary artery disease | 1014 | 20120 | 2.7E-08 | 1.312 | [1.192,1.444] |
| Heart attack | 486 | 22498 | 3.9E-05 | 1.318 | [1.155,1.503] |
| High blood pressure | 5230 | 15797 | 0.0091 | 1.065 | [1.016,1.117] |
| High cholesterol | 7970 | 15030 | 0.18 | 1.029 | [0.987,1.073] |
Concluding thoughts
What might explain the failure to replicate The Lancet finding in the 23andMe cohort? One possibility is that it is simply a false positive, and the apparent replication in the original study was just luck. Another possibility is that the association is real but restricted to the original study population of British men. That is, it might replicate in a new study of British men, but might not generalize well. Distinguishing between these possibilities will probably require additional studies, since it is very difficult to “prove a negative” and establish that a hypothetical association does not exist. The authors suggest that their study is the first to evaluate association between Y chromosome haplogroups and CAD. However, rs2032597 is present on recent Illumina SNP arrays used for GWAS, so data for this variant should exist in other large studies of CAD. Thus, it should be possible to quickly test many cohorts for this association, even if the Y chromosome data has not been previously analyzed.