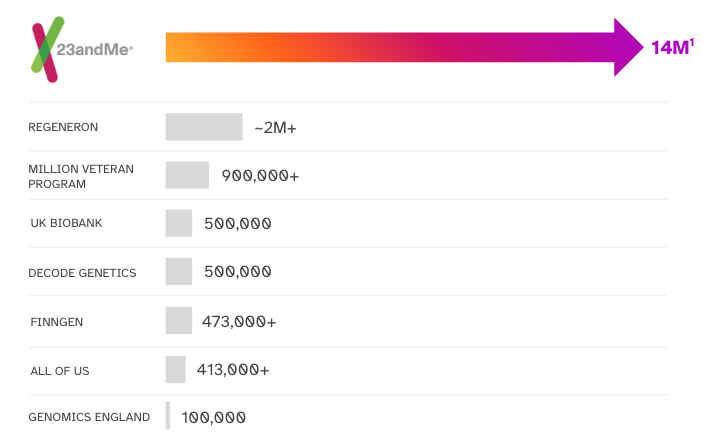
Analyses of large genotyped and phenotyped datasets can provide valuable information for therapeutic development and research. Many sizable genetic research data sets have been used for this purpose, such as the UK Biobank, the Million Veterans Project, and the NIH’s All of Us.
What is unique about 23andMe’s approach is that we have data at a scale that significantly exceeds many other entities. The size of our data increases our ability to detect disease associations with a given genotype and identify individuals with unique genomic composition for participation in additional research opportunities.
Early results from a pilot project 23andMe launched this year inspired us to look deeper into our Research Cohort at the gene MAP3K15, where previous research found a loss of function variant that protects people against developing type 2 diabetes.
If someone has a loss of function variant, for example, the MAP3K15 loss of function variant, and they are healthy, it tells scientists that turning off that gene could be safe. Knowing that a loss of function variant is not deleterious is incredibly important for drug development. Understanding this helps not just with target validation but with potentially de-risking and safety concerns around developing therapeutics that might turn off the expression of that gene.
Unique Insights into MAP3K15
Our researchers wanted to see if the size of the 23andMe database could enable additional insight into the metabolic role of MAP3K15 that might not have been discoverable in other datasets. The findings from this work was recently published on the preprint server MedRxiv.
For this study we performed various genetic analyses for MAP3K15 to see what we could learn from the >11 million consented customers in the 23andMe Research Cohort. In addition to confirming the protective effect for diabetes, analysis of our database resulted in several new insights for MAP3K15 loss of function (LoF):
- MAP3K15 LoF is associated with an increased risk of high blood pressure
- MAP3K15 LoF is associated with a decreased risk of high cholesterol
- Among individuals determined to be at a high genetic risk for type 2 diabetes, those who also have MAP3K15 LoF show a potential 4.5-year delay in the median age of disease onset compared to non-carriers
Genetics-driven Recruitment
We wanted to know if we could establish a recruitment capability to obtain clinical laboratory evidence for the new MAP3K15 findings from genetic analyses of the 23andMe database. Because we have a very, very large number of people who have consented to participate in this additional level of research, we could readily identify potential research participants of interest with the protective MAP3K15 loss of function variant.
In this study, we were particularly interested in clinical lab values related to participant blood pressure and high cholesterol in individuals with or without a particular loss of function variant in MAP3K15. To obtain these lab values and biobank blood samples from consented participants, we set up a research protocol to recruit participants nationwide to answer survey questions and participate in a blood draw. This protocol was reviewed and approved by an Institutional Review Board (an independent ethical and regulatory review board) to ensure protection for the rights and welfare of our study participants.
Results from this pilot study enabled us to find evidence supporting our database findings, including elevated systolic blood pressure, reduced cholesterol levels, and lower LDL/HDL ratio in 23andMe participants with the protective variant compared to controls without the variant.
The participants in this study represent a group of our research participants who’ve gone through an additional layer of consent.
For this research, our research participants have opted-in to being contacted by our researchers about research opportunities based on genetic information not included in their 23andMe reports.
These research opportunities might be related to specific conditions for which we do not yet have a report. Those who agree to participate and who are contacted for specific studies may only learn about the condition being studied but not the specific variant.
Like all our research, participants must opt in to be included, and those currently participating can change their minds and then opt out at any time.
Learn more here.
DNA-Driven Discoveries
This recruitment capability opens a whole host of capabilities for researchers. Dubbed “The DNA-Driven Discoveries Study,” this genetics-driven recruitment approach could potentially inform drug target safety concerns, improve patient selection and efficacy in clinical trials while also helping to identify hard-to-find drug targets, rare genetic variants and even reveal more about the underlying biology of certain conditions.
“I’m not aware of any other entity out there with this capability and scale,” said Jen Brady, Ph.D., a senior scientist in 23andMe’s Therapeutics division who helped lead the study. “We’re leveraging the discovery power of what is the world’s largest re-contactable genetic database.
Our (DNA-Driven Discoveries) study allows 23andMe scientists to recontact consented individuals based on genetic variants they may have but do not know about. Contacting these individuals based on their genetics is a key capability for 23andMe. It could help inform drug discovery efforts for 23andMe and our collaborators, improve how we create genetic research hypotheses, and help us build an internal deeply-phenotyped biobank.”
Next Steps
We believe the ability to identify, recruit, and characterize research participants based on genetics unknown to them is a critical capability for 23andMe to inform drug development and genetics research.
Future studies using this method could enable similar recruitment and study designs for other conditions. They could also potentially help in clinical trials as we determine a drug’s efficacy and potential toxicity, modeling control and treatment arms. It may also help recruit family members, which could increase power for studying, especially rare variants that might run in families. We may also use this genetic-driven research to test the validity or interpretation of some self-reported data from research participants.
These DNA-driven discoveries will improve our important work and may lead to important insights into the treatment and diagnosis of disease.