A segment of chromosome 14 folded to reveal a fractal curve using Origami. Designed and folded by Jason Ku. Photo by Erik Demaine.
A segment of chromosome 14 folded to reveal a fractal curve using Origami. Designed and folded by Jason Ku. Photo by Erik Demaine.
How do you get three billion pairs of As, Cs, Ts and Gs–about six feet worth of DNA–into the nucleus of a tiny cell?
Most students of biology would answer by saying that this is accomplished by tightly coiling up the DNA.
Oh yeah?
Well, how is it coiled?
Tighten Up
As cells perform different functions and respond to different environmental signals, proteins that help turn genes on and off need to quickly gain access to different parts of the genome. That means DNA needs to be arranged in such a way that it won’t get all tangled up.
Packing DNA like luggage at the end of a vacation, with everything smashed together and shoved in any which way, just won’t cut it.
Hi-C
Using a new technique called “Hi-C,” researchers at Harvard, MIT and the University of Massachusetts appear to have solved the riddle. Their results, published today in the journal Science, show that nature has devised quite an elegant storage solution.
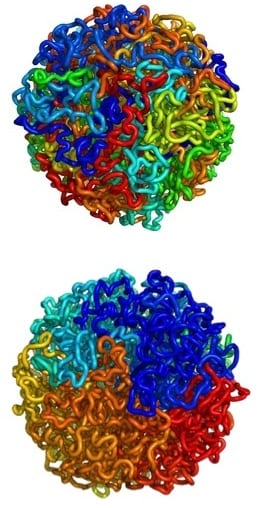 Equilibrium (top) and fractal (bottom) globules. Nearby regions on a chain of DNA are indicated using similar colors. The equilibrium globule is highly entangled; regions nearby along the chain are far apart in 3D. In the fractal globule, regions nearby along the chain are also nearby in 3D. Images: Leonid A. Mirny and Maxim Imakaev
Equilibrium (top) and fractal (bottom) globules. Nearby regions on a chain of DNA are indicated using similar colors. The equilibrium globule is highly entangled; regions nearby along the chain are far apart in 3D. In the fractal globule, regions nearby along the chain are also nearby in 3D. Images: Leonid A. Mirny and Maxim Imakaev
The scientists first treated cells with formaldehyde to freeze the DNA in place. They then used enzymes to break the DNA apart and put it back together in a different configuration.
A final step of sequencing allowed them to identify pieces of DNA that are naturally close together in the nucleus.
“We made a fantastic three-dimensional jigsaw puzzle and then, with a computer, solved the puzzle,” said co-first author Nynke van Berkum in a statement.
Two important aspects of DNA organization emerged. First, there are two main compartments in the nucleus — one for DNA that is in use and one that acts as a storage facility for unneeded sequences.
“Cells cleverly separate the most active genes into their own special neighborhood, to make it easier for proteins and other regulators to reach them,” said one of the paper’s senior authors, Job Dekker of UMass Medical School, in a statement.
Globule of Globules
The other striking aspect of the nucleus is that chromosomes appear to be folded up into an arrangement called a fractal globule, which the authors described as a “beads-on-a-string” configuration. Multiple rounds of “crumpling” of the DNA into beads leads to a “globule-of-globules-of-globules.”
Previous models suggested that DNA was in a more random arrangement called an equilibrium globule. This configuration, however, is known to be prone to dense knotting. Fractal globules are knot-free.
Image Source: Harvard School of Engineering and Applied Sciences